ChronoRoot 2.0
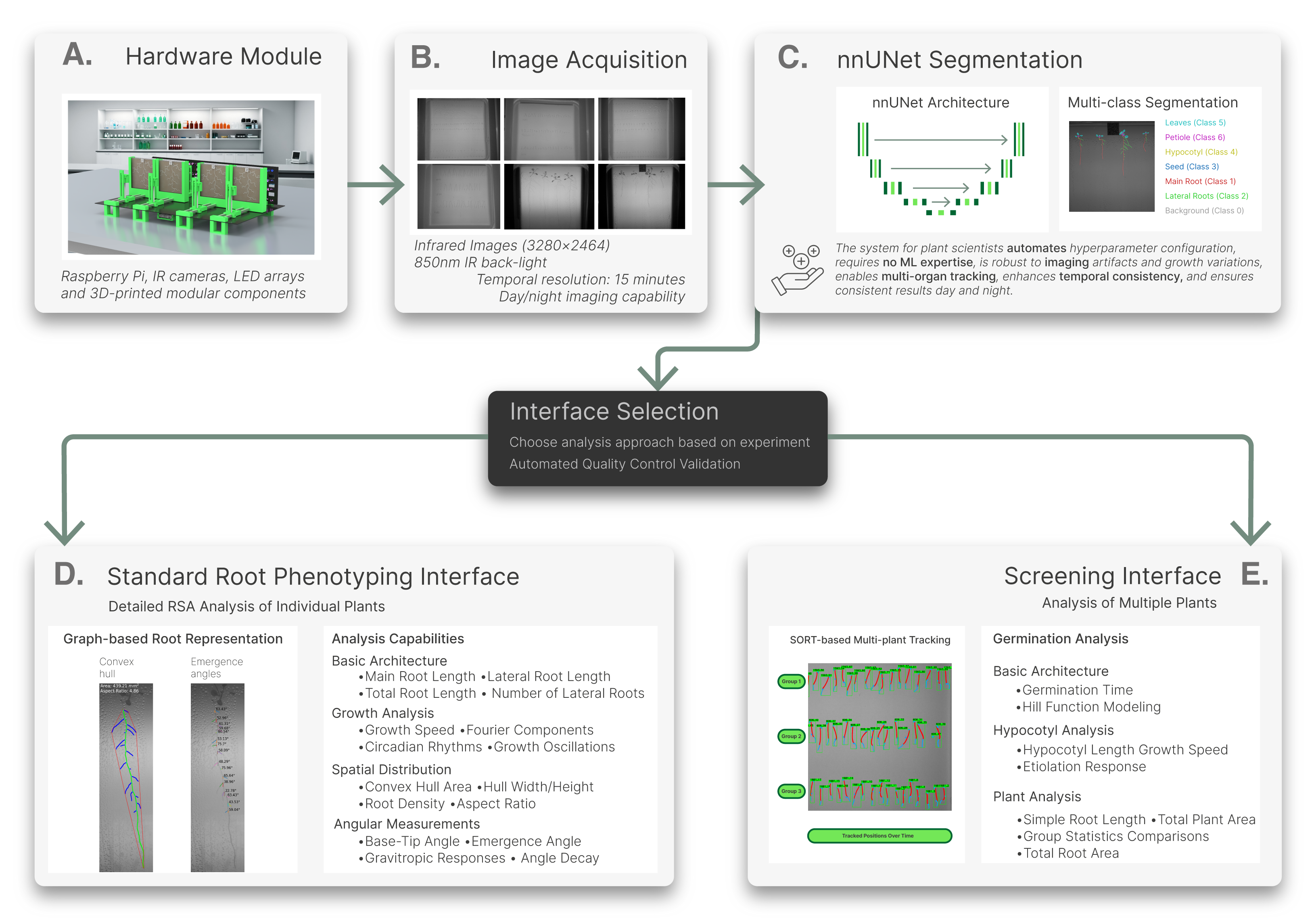
ChronoRoot 2.0 is a comprehensive, open-source solution that merges affordable hardware with state-of-the-art Deep Learning to automate the tracking and analysis of plant development. It includes three specialized graphical interfaces. On Linux and Windows 10 (via WSL2), these are installed with custom desktop launchers for a point-and-click experience.
| Icon | Application | Purpose & Capabilities | Tutorials |
|---|---|---|---|
| Segmentation App | Multi-class segmentation tool: Uses the nnU-Net engine to segment 6 plant organs. | Segmentation Guide - Training Guide | |
| Standard Interface | Detailed RSA Analysis: Graph-based analysis of individual plants. Measures growth speeds, gravitropic angles, and different RSA metrics. | Arabidopsis Guide - Tomato Guide | |
| Screening Interface | High-throughput screening: Automated tracking for up to 100 seeds/plate. Optimized for germination and etiolation studies. | Germination Guide - Etiolation Guide |
Choosing your workflow
All experiments begin in the Segmentation App to generate segmentation masks. From there, choose the Standard Interface if you require detailed root architecture (4-6 plants/plate) or the Screening Interface for high-density populations (up to 100 seeds/plate).
Latest News & Major Updates
- 28 Jan 2026: Hugging Face Integration: Model weights are now hosted on the Hugging Face Hub!
- The repository is now lightweight and clone-friendly.
- Installers have been updated to fetch the latest models automatically.
- Added a download weights script for easy manual updates.
- 07 Jan 2026: Added desktop shortcut support for Windows 10 users (via WSL2).
- 15 Dec 2025: Updated desktop launchers with custom ChronoRoot icons.
- 11 Dec 2025: Introduced Apptainer as the primary installer to support HPC clusters. Docker is now reserved for Mac or advanced users.
- 08 Nov 2025: Unified the repository into a single Conda environment for simplified maintenance and setup.
- 30 Oct 2025: nnUNetv2 Port:
- Added a new CLI and removed the dependency on the custom nnUNet fork; models now load directly via PyTorch.
- New model weights are available for Arabidopsis thaliana and Tomato multi-class segmentation.
- 20 Apr 2025: 🚀 ChronoRoot 2.0 Released! Featuring a revamped segmentation engine, enhanced analysis modules, and improved UX.
Training Dataset Changelog
All updates refer to the ChronoRoot Dataset on HuggingFace.
- 07 Nov 2025: Added 48 Arabidopsis thaliana images (hypocotyl etiolation), bringing the species total to 913.
- 30 Oct 2025: Major expansion and restructuring:
- Added 480 Solanum lycopersicum (tomato) images with multi-class masks.
- Added 170 Arabidopsis thaliana images (for germination and multiple plants root analysis), totaling 863.
- Reorganized directory structure to match biologist annotation folders.
- Removed legacy nnUNet checkpoints to optimize storage following the ChronoRoot 2.0 deployment.
- 30 Jan 2025: Converted 526 existing masks from binary to multi-class. Added 167 new images (germination and etiolation), totaling 693.
- 30 Apr 2024: Initial release of 526 Arabidopsis thaliana root images with binary segmentation masks.
Installation
Choose the method that best fits your setup. Native and Apptainer installations provide three desktop shortcuts for ease of use.
Automatic Model Download
The installers below will automatically check for and download the latest segmentation model weights from Hugging Face during setup.
Windows 11 Users
Note, for Windows 11 users shortcuts are not yet available. Please run ChronoRoot applications via WSL terminal, we will be adding shortcut support in a near future update.
Best for: Linux or Windows 10 WSL where you want desktop integration. Requires Miniconda or Anaconda installed in your system.
Ubuntu / Linux:
wget https://raw.githubusercontent.com/chronoroot/ChronoRoot2/master/installer_conda_linux.sh
bash installer_conda_linux.sh
Windows 10 (via WSL2):
wget https://raw.githubusercontent.com/chronoroot/ChronoRoot2/master/installer_conda_wsl.sh
bash installer_conda_wsl.sh
Best for: HPC Clusters or single-file container users.
Linux:
wget https://raw.githubusercontent.com/chronoroot/ChronoRoot2/master/apptainerInstaller/installer_linux.sh
bash installer_linux.sh
Windows 10 (via WSL2):
wget https://raw.githubusercontent.com/chronoroot/ChronoRoot2/master/apptainerInstaller/installer_windows.sh
bash installer_windows.sh
Best for: macOS users or advanced sandbox environments.
docker pull ngaggion/chronoroot:latest
System Requirements
- RAM: 8 GB min (16 GB recommended)
- GPU: NVIDIA GPU with CUDA 11.0+ (Highly recommended for Segmentation)
- macOS Note: Native installation is not supported; use Docker.
Demo Data and Tutorials
New to ChronoRoot? Download our Demo Data Videos and follow these guides to process your first dataset:
- Root Analysis – Arabidopsis thaliana (Standard Interface)
- Root Analysis – Tomato (Standard Interface)
- Germination Analysis – Arabidopsis thaliana (Screening Interface)
- Hypocotyl Etiolation Analysis – Arabidopsis thaliana (Screening Interface)
Citation
If you use ChronoRoot 2.0 in your research, please cite our work:
@article{gaggion2025chronoroot,
title={ChronoRoot 2.0: An Open AI-Powered Platform for 2D Temporal Plant Phenotyping},
author={Gaggion, Nicolás et al.},
journal={arXiv preprint arXiv:2504.14736},
year={2025}
}
Centralized Resources
- Software Repository: https://github.com/ChronoRoot/ChronoRoot2
- Hardware Module: https://github.com/ChronoRoot/ChronoRootModuleHardware
- Module Control: https://github.com/ChronoRoot/ChronoRootControl
- Training Dataset: https://huggingface.co/datasets/ngaggion/ChronoRoot2
- Demo Data: https://drive.google.com/drive/folders/1PJCn_MMHcM9KPgz8dYe1F2Cvdt43FS3Z?usp=sharing
Model Weights: https://huggingface.co/ngaggion/models



